Task 4: Cell segmentation

Description
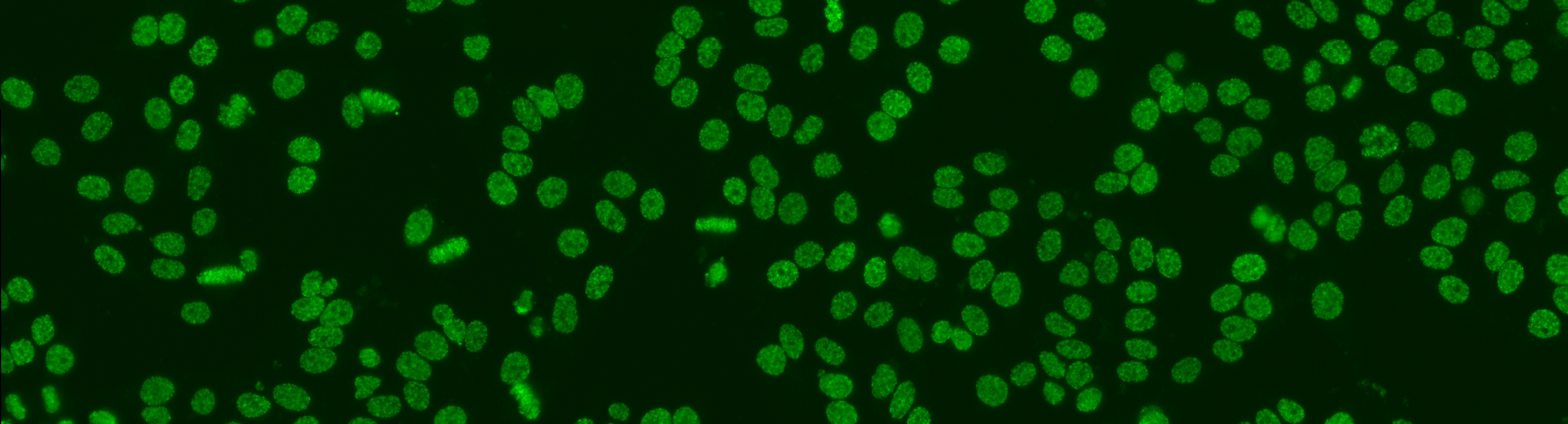
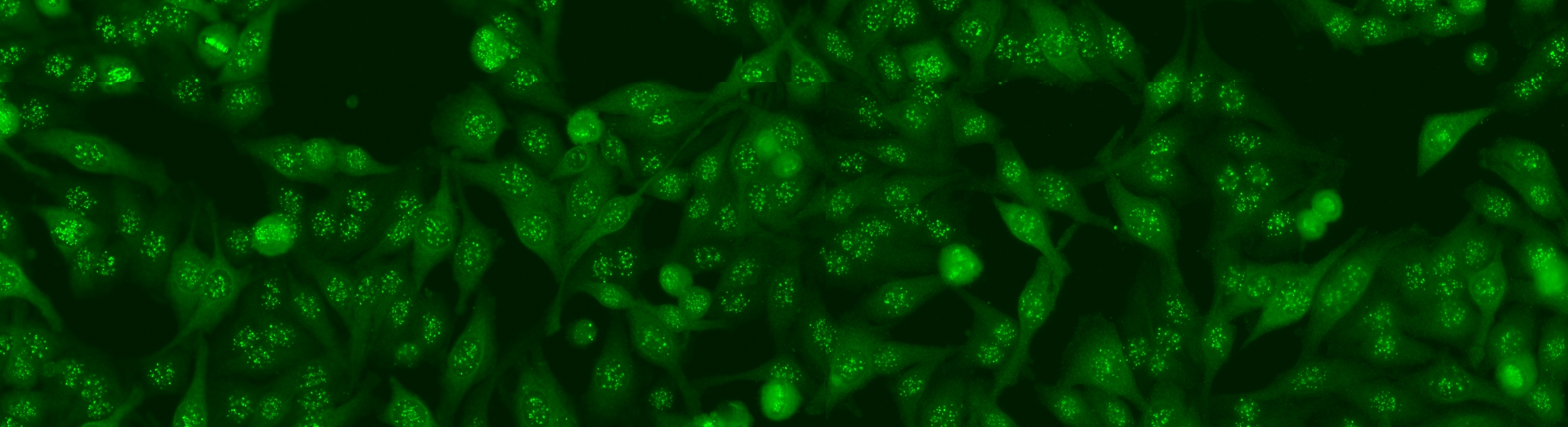
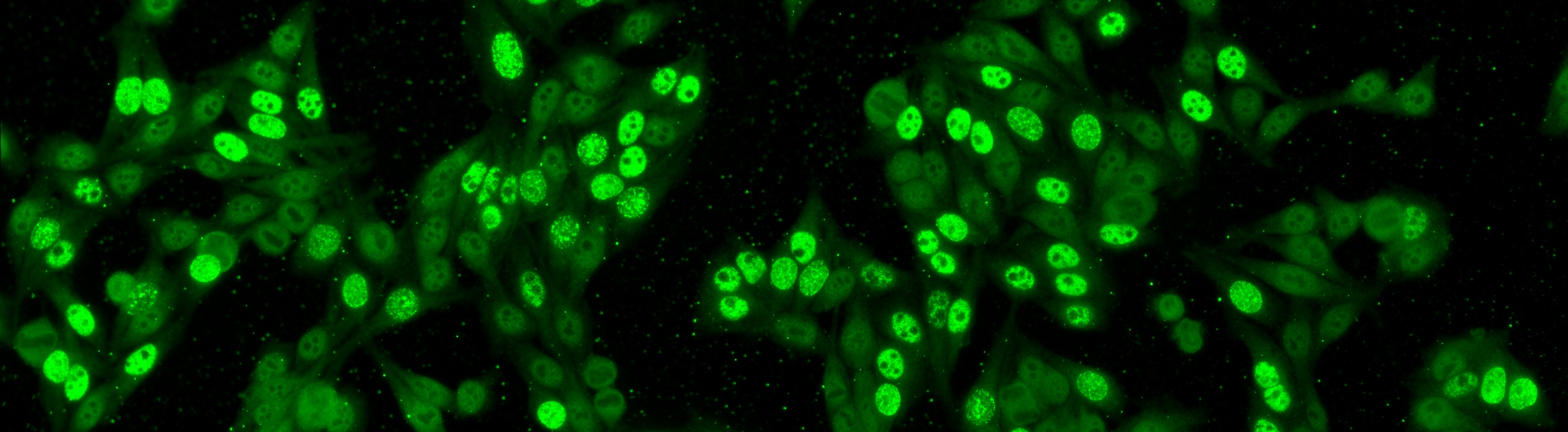
Cell segmentation is the first step on the whole analysis. Despite its high difficulties, the segmentation problem can be solved by the secondary channel using DAPI. Nevertheless, the introduction of DAPI may complicate the laboratory work flow; thus, may not even practical in some pathology laboratories. This means, the cell segmentation on the primary channel is still not solved yet. The aim for this task is using only the primary channel, to generate the segmentation map that is equivalent to the map generated from the secondary channel.
Dataset
As the segmentation map from the DAPI channel can be considered as the gold standard, we opt to use the images acquired for the second task. This mean the dataset will consist of 1,001 specimen images. For each image we provide both the original image and the segmentation mask obtained from the DAPI channel; the segmentation mask will be used as the ground truth with respect the performance will be evaluated.
Evaluation
The assessment of the segmentation quality has been carried out according to the procedure described in [18] in order to report the performance at the cell level. The metric that will be adopted for reporting performance of each method is the f-index calculated as:

where:
![]()
with:
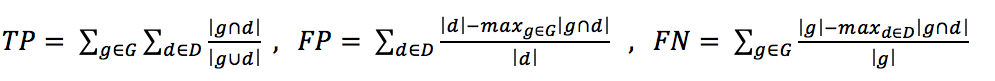
and D and G are the sets of the detected cells and of the ground-truth cells, respectively, while | · | denotes the cardinality of a set.