Task 2: Specimen level pattern classification

Description
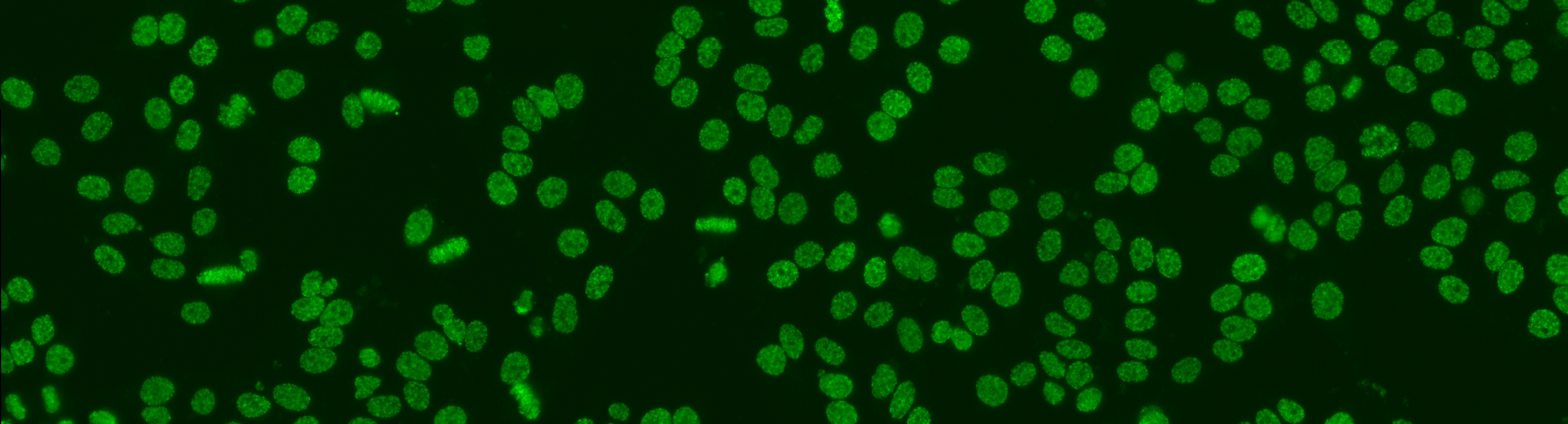
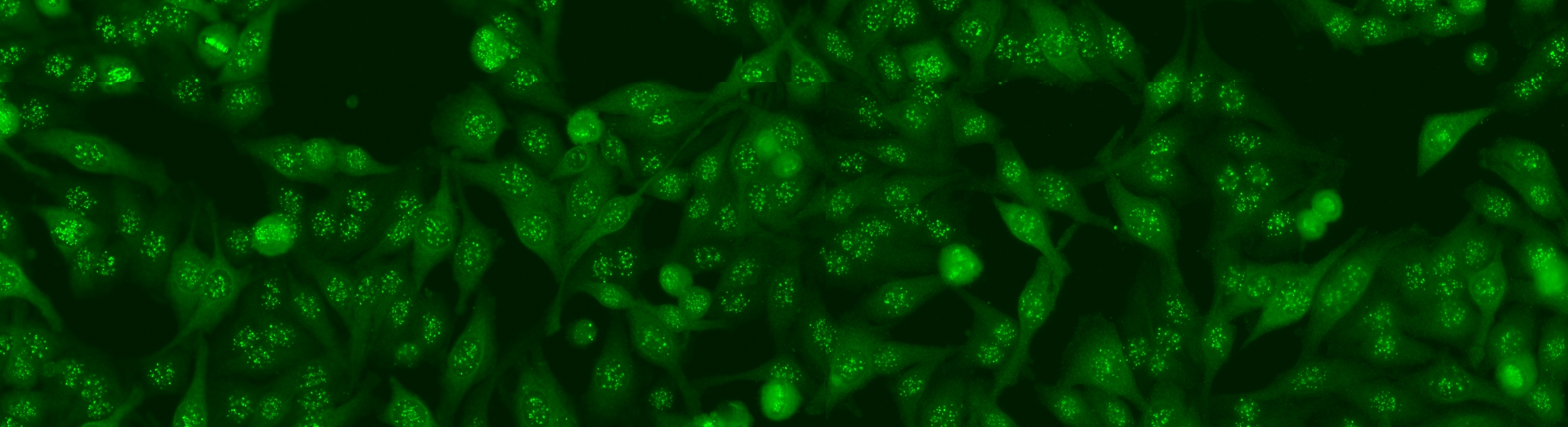
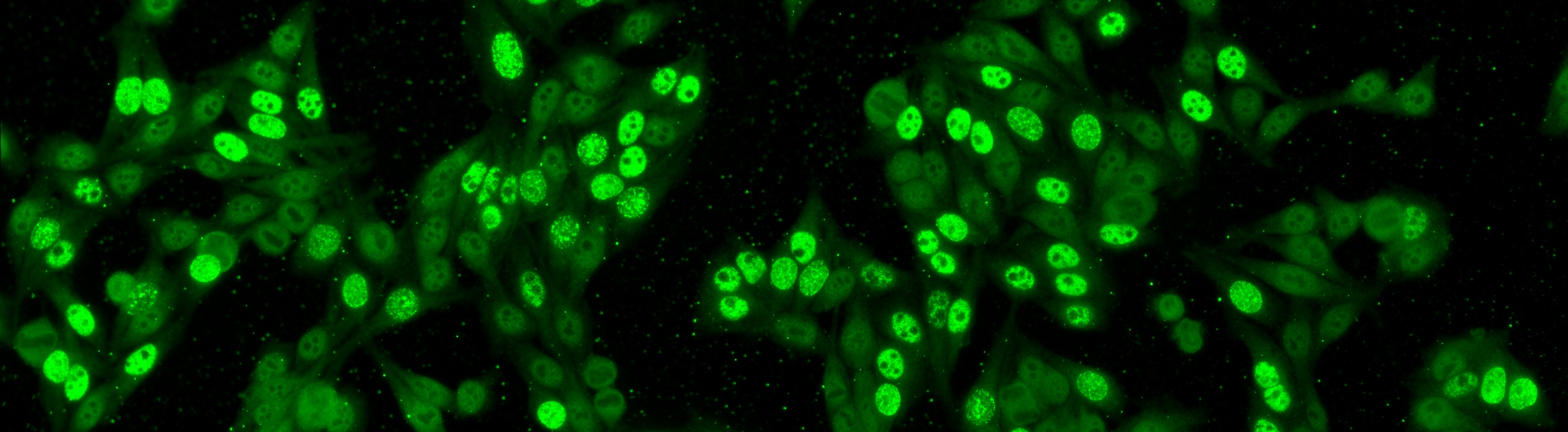
The aim of the task is to classify the patient specimen. Each patient specimen (refer to Fig. 4) have multiple HEp-2 cells. This task will equip the participants with the segmented HEp-2 cells although the mitotic cells are not indicated. The participants have the liberty to either work directly on the specimen images (i.e. without using the segmentation map), or utilize the provided segmentation map. This task has been firstly proposed at the ICPR 2014 competition and similarly to the cell-level pattern classification task. Again, the task will be the continuation from the previous year to witness the advances made by the community.
Dataset
The dataset was acquired in 2013 at Sullivan Nicolaides Pathology Laboratory, Australia. It was collected from 1001 patient sera with positive ANA test. Each patient sera were diluted to 1:80 and the specimen was photographed using a monochrome camera fitted on a microscope. Each specimen was photographed at four different locations rendering each specimen has four images. The dataset has seven pattern classes: homogeneous, speckled, nucleolar, centromere, nuclear membrane, golgi and mitotic spindle. The first four classes represent common ANA patterns whilst the rest three classes are less common.
The dataset is divided into training and test set as following: approximately ¼ sera in the training set and the remaining in the test set. All images are in monochromatic uncompressed format with resolution 1388 x 1040 pixels together with their corresponding cell mask which was obtained automatically.
The labelling process has involved at least two scientists who read each patient specimen under a microscope. A third expert’s opinion has been sought to adjudicate any discrepancy between the two opinions. Each slide image is also provided with the corresponding class (one among the patterns defined above). Furthermore, all the labels were validated by using secondary tests such as ENA and anti-ds-DNA in order to confirm the presence and/absence of specific patterns.
Each specimen image contained in the database is annotated with the following information:
- Staining pattern of the specimen
- Intensity of the specimen
- Mask of the specimen
Evaluation
The application obtaining the highest value of the mean class accuracy (MCA) in specimen classification over the test set will be declared as the winner. The proclamation of the winner will be made during the contest session at ICPR 2016.